Note
Go to the end to download the full example code
Dataframe as an input#
A pipeline usually ingests data as a matrix. It may be converted in a matrix if all the data share the same type. But data held in a dataframe have usually multiple types, float, integer or string for categories. ONNX also supports that case.
A dataset with categories#
from mlinsights.plotting import pipeline2dot
import numpy
import pprint
from onnx.reference import ReferenceEvaluator
from onnxruntime import InferenceSession
from pandas import DataFrame
from sklearn.pipeline import Pipeline
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import OneHotEncoder
from sklearn.ensemble import RandomForestClassifier
from pyquickhelper.helpgen.graphviz_helper import plot_graphviz
from skl2onnx import to_onnx
from skl2onnx.algebra.type_helper import guess_initial_types
data = DataFrame(
[
dict(CAT1="a", CAT2="c", num1=0.5, num2=0.6, y=0),
dict(CAT1="b", CAT2="d", num1=0.4, num2=0.8, y=1),
dict(CAT1="a", CAT2="d", num1=0.5, num2=0.56, y=0),
dict(CAT1="a", CAT2="d", num1=0.55, num2=0.56, y=1),
dict(CAT1="a", CAT2="c", num1=0.35, num2=0.86, y=0),
dict(CAT1="a", CAT2="c", num1=0.5, num2=0.68, y=1),
]
)
cat_cols = ["CAT1", "CAT2"]
train_data = data.drop("y", axis=1)
categorical_transformer = Pipeline(
[("onehot", OneHotEncoder(sparse=False, handle_unknown="ignore"))]
)
preprocessor = ColumnTransformer(
transformers=[("cat", categorical_transformer, cat_cols)], remainder="passthrough"
)
pipe = Pipeline([("preprocess", preprocessor), ("rf", RandomForestClassifier())])
pipe.fit(train_data, data["y"])
Display.
dot = pipeline2dot(pipe, train_data)
ax = plot_graphviz(dot)
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
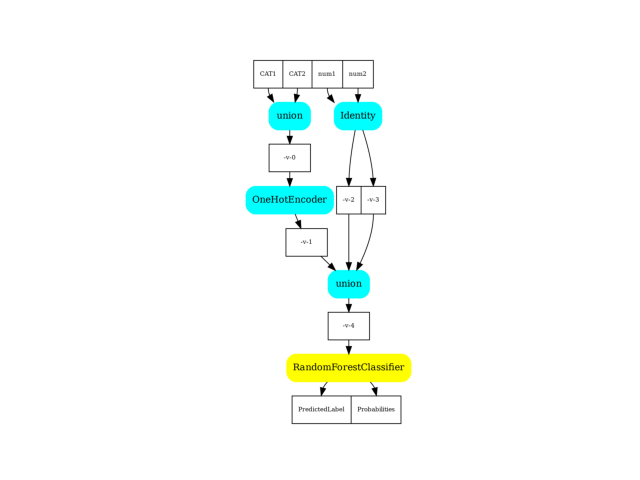
Conversion to ONNX#
Function to_onnx does not handle dataframes.
onx = to_onnx(pipe, train_data[:1], options={RandomForestClassifier: {"zipmap": False}})
Prediction with ONNX#
onnxruntime does not support dataframes.
sess = InferenceSession(onx.SerializeToString(), providers=["CPUExecutionProvider"])
try:
sess.run(None, train_data)
except Exception as e:
print(e)
run(): incompatible function arguments. The following argument types are supported:
1. (self: onnxruntime.capi.onnxruntime_pybind11_state.InferenceSession, arg0: List[str], arg1: Dict[str, object], arg2: onnxruntime.capi.onnxruntime_pybind11_state.RunOptions) -> List[object]
Invoked with: <onnxruntime.capi.onnxruntime_pybind11_state.InferenceSession object at 0x7f4512d02cf0>, ['label', 'probabilities'], CAT1 CAT2 num1 num2
0 a c 0.50 0.60
1 b d 0.40 0.80
2 a d 0.50 0.56
3 a d 0.55 0.56
4 a c 0.35 0.86
5 a c 0.50 0.68, None
Let’s use a shortcut
oinf = ReferenceEvaluator(onx)
got = oinf.run(None, train_data)
print(pipe.predict(train_data))
print(got["label"])
Traceback (most recent call last):
File "/home/xadupre/github/sklearn-onnx/docs/tutorial/plot_gbegin_dataframe.py", line 91, in <module>
got = oinf.run(None, train_data)
File "/home/xadupre/github/onnx/onnx/reference/reference_evaluator.py", line 526, in run
outputs = node.run(*inputs, **linked_attributes)
File "/home/xadupre/github/onnx/onnx/reference/op_run.py", line 477, in run
raise TypeError(
TypeError: Issues with types [<class 'pandas.core.series.Series'>, <class 'pandas.core.series.Series'>] and attributes ['axis'] and linked attributes=[] (operator 'Concat').
And probilities.
print(pipe.predict_proba(train_data))
print(got["probabilities"])
It looks ok. Let’s dig into the details to directly use onnxruntime.
Unhide conversion logic with a dataframe#
A dataframe can be seen as a set of columns with different types. That’s what ONNX should see: a list of inputs, the input name is the column name, the input type is the column type.
def guess_schema_from_data(X):
init = guess_initial_types(X)
unique = set()
for _, col in init:
if len(col.shape) != 2:
return init
if col.shape[0] is not None:
return init
if len(unique) > 0 and col.__class__ not in unique:
return init
unique.add(col.__class__)
unique = list(unique)
return [("X", unique[0]([None, sum(_[1].shape[1] for _ in init)]))]
init = guess_schema_from_data(train_data)
pprint.pprint(init)
Let’s use float instead.
for c in train_data.columns:
if c not in cat_cols:
train_data[c] = train_data[c].astype(numpy.float32)
init = guess_schema_from_data(train_data)
pprint.pprint(init)
Let’s convert with skl2onnx only.
onx2 = to_onnx(
pipe, initial_types=init, options={RandomForestClassifier: {"zipmap": False}}
)
Let’s run it with onnxruntime. We need to convert the dataframe into a dictionary where column names become keys, and column values become values.
inputs = {c: train_data[c].values.reshape((-1, 1)) for c in train_data.columns}
pprint.pprint(inputs)
Inference.
sess2 = InferenceSession(onx2.SerializeToString(), providers=["CPUExecutionProvider"])
got2 = sess2.run(None, inputs)
print(pipe.predict(train_data))
print(got2[0])
And probilities.
print(pipe.predict_proba(train_data))
print(got2[1])
Total running time of the script: (0 minutes 0.602 seconds)