Note
Go to the end to download the full example code
Converter for WOE#
WOE means Weights of Evidence. It consists in checking that a feature X belongs to a series of regions - intervals -. The results is the label of every intervals containing the feature.
A simple example#
X is a vector made of the first ten integers. Class
WOETransformer
checks that every of them belongs to two intervals,
]1, 3[ (leftright-opened) and [5, 7]
(left-right-closed). The first interval is associated
to weight 55 and and the second one to 107.
import os
import numpy as np
import pandas as pd
from onnx.tools.net_drawer import GetPydotGraph, GetOpNodeProducer
from onnxruntime import InferenceSession
import matplotlib.pyplot as plt
from skl2onnx import to_onnx
from skl2onnx.sklapi import WOETransformer
# automatically registers the converter for WOETransformer
import skl2onnx.sklapi.register # noqa
X = np.arange(10).astype(np.float32).reshape((-1, 1))
intervals = [[(1.0, 3.0, False, False), (5.0, 7.0, True, True)]]
weights = [[55, 107]]
woe1 = WOETransformer(intervals, onehot=False, weights=weights)
woe1.fit(X)
prd = woe1.transform(X)
df = pd.DataFrame({"X": X.ravel(), "woe": prd.ravel()})
df
One Hot#
The transformer outputs one column with the weights. But it could return one column per interval.
woe2 = WOETransformer(intervals, onehot=True, weights=weights)
woe2.fit(X)
prd = woe2.transform(X)
df = pd.DataFrame(prd)
df.columns = ["I1", "I2"]
df["X"] = X
df
In that case, weights can be omitted. The output is binary.
woe = WOETransformer(intervals, onehot=True)
woe.fit(X)
prd = woe.transform(X)
df = pd.DataFrame(prd)
df.columns = ["I1", "I2"]
df["X"] = X
df
Conversion to ONNX#
skl2onnx implements a converter for all cases.
onehot=False
onx1 = to_onnx(woe1, X)
sess = InferenceSession(onx1.SerializeToString(), providers=["CPUExecutionProvider"])
print(sess.run(None, {"X": X})[0])
[[ 0.]
[ 0.]
[ 55.]
[ 0.]
[ 0.]
[107.]
[107.]
[107.]
[ 0.]
[ 0.]]
onehot=True
onx2 = to_onnx(woe2, X)
sess = InferenceSession(onx2.SerializeToString(), providers=["CPUExecutionProvider"])
print(sess.run(None, {"X": X})[0])
[[ 0. 0.]
[ 0. 0.]
[ 55. 0.]
[ 0. 0.]
[ 0. 0.]
[ 0. 107.]
[ 0. 107.]
[ 0. 107.]
[ 0. 0.]
[ 0. 0.]]
ONNX Graphs#
onehot=False
pydot_graph = GetPydotGraph(
onx1.graph,
name=onx1.graph.name,
rankdir="TB",
node_producer=GetOpNodeProducer(
"docstring", color="yellow", fillcolor="yellow", style="filled"
),
)
pydot_graph.write_dot("woe1.dot")
os.system("dot -O -Gdpi=300 -Tpng woe1.dot")
image = plt.imread("woe1.dot.png")
fig, ax = plt.subplots(figsize=(10, 10))
ax.imshow(image)
ax.axis("off")
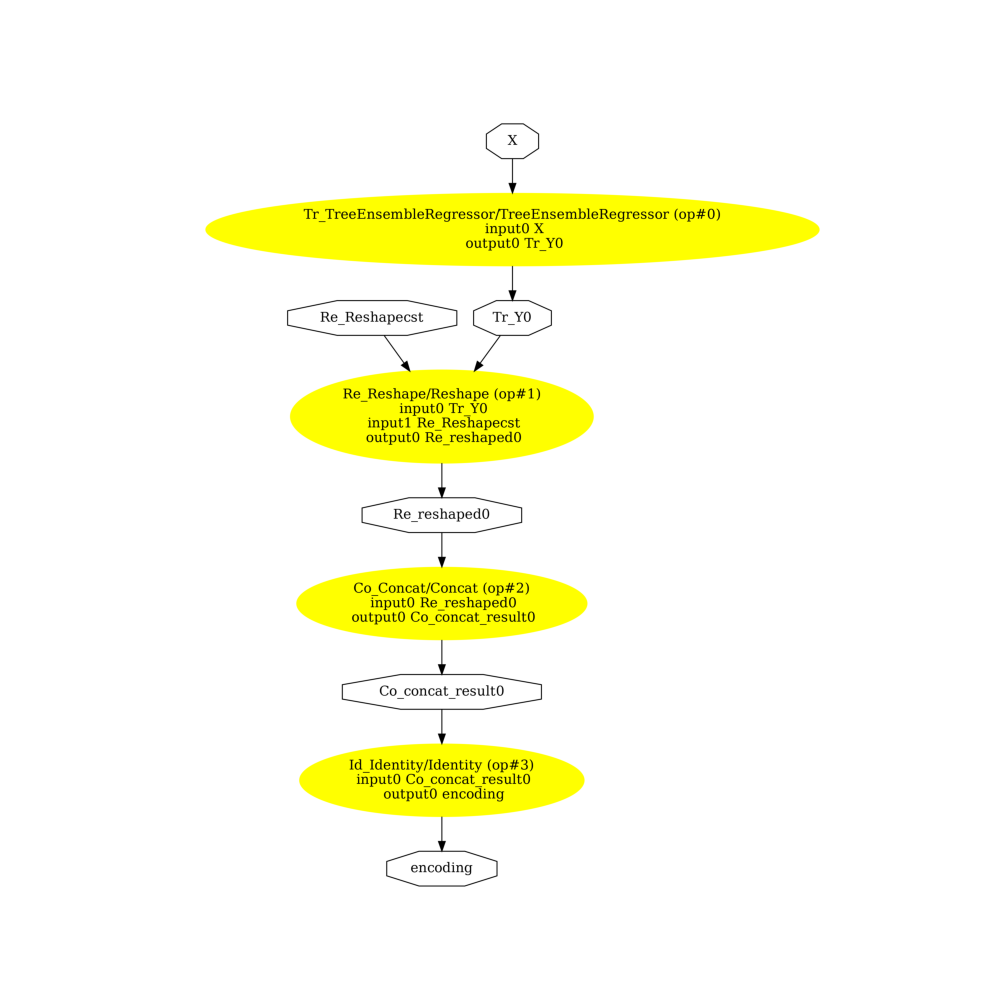
(-0.5, 2674.5, 3321.5, -0.5)
onehot=True
pydot_graph = GetPydotGraph(
onx2.graph,
name=onx2.graph.name,
rankdir="TB",
node_producer=GetOpNodeProducer(
"docstring", color="yellow", fillcolor="yellow", style="filled"
),
)
pydot_graph.write_dot("woe2.dot")
os.system("dot -O -Gdpi=300 -Tpng woe2.dot")
image = plt.imread("woe2.dot.png")
fig, ax = plt.subplots(figsize=(10, 10))
ax.imshow(image)
ax.axis("off")
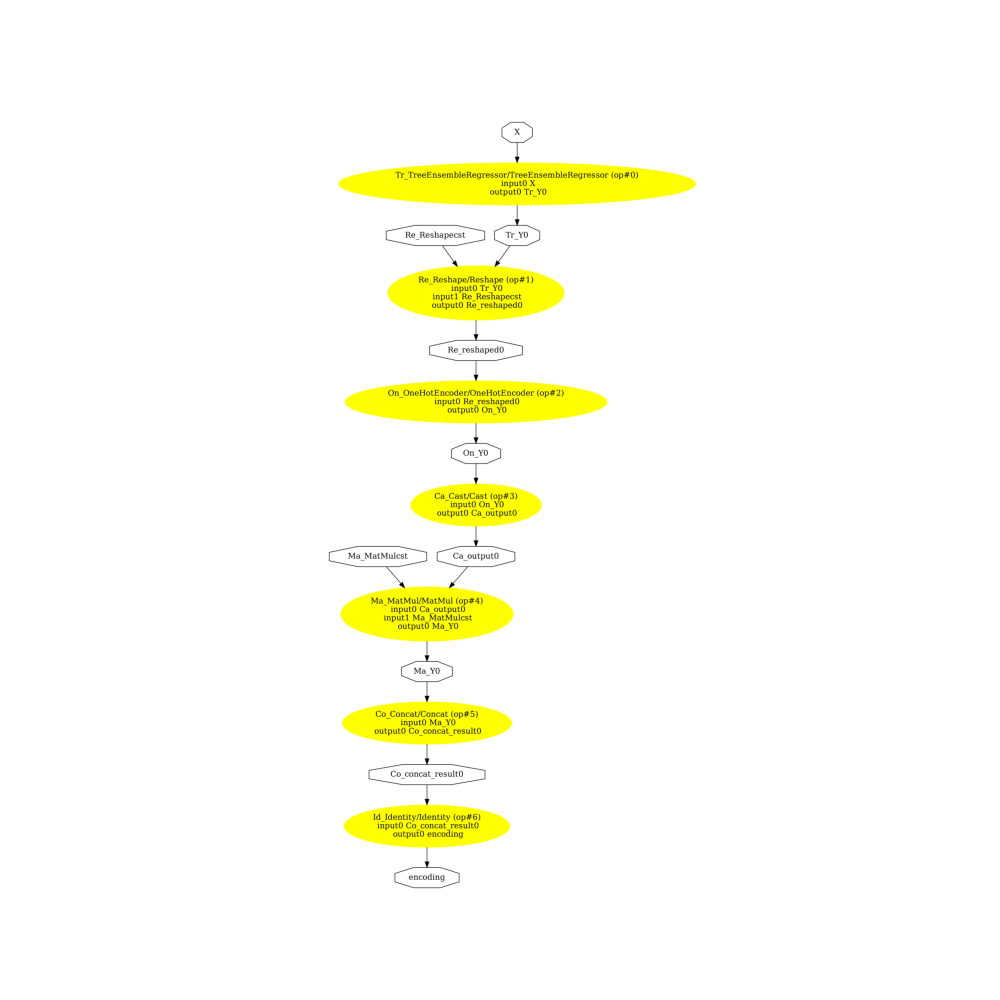
(-0.5, 2743.5, 5696.5, -0.5)
Half-line#
An interval may have only one extremity defined and the other can be infinite.
intervals = [[(-np.inf, 3.0, True, True), (5.0, np.inf, True, True)]]
weights = [[55, 107]]
woe1 = WOETransformer(intervals, onehot=False, weights=weights)
woe1.fit(X)
prd = woe1.transform(X)
df = pd.DataFrame({"X": X.ravel(), "woe": prd.ravel()})
df
And the conversion to ONNX using the same instruction.
onxinf = to_onnx(woe1, X)
sess = InferenceSession(onxinf.SerializeToString(), providers=["CPUExecutionProvider"])
print(sess.run(None, {"X": X})[0])
[[ 55.]
[ 55.]
[ 55.]
[ 55.]
[ 0.]
[107.]
[107.]
[107.]
[107.]
[107.]]
Total running time of the script: (0 minutes 3.955 seconds)