Note
Go to the end to download the full example code.
Convert a model with a reduced list of operators¶
Some runtime dedicated to onnx do not implement all the operators and a converted model may not run if one of them is missing from the list of available operators. Some converters may convert a model in different ways if the users wants to blacklist some operators.
GaussianMixture¶
The first converter to change its behaviour depending on a black list of operators is for model GaussianMixture.
import onnxruntime
import onnx
import numpy
import os
from timeit import timeit
import numpy as np
import matplotlib.pyplot as plt
from onnx.tools.net_drawer import GetPydotGraph, GetOpNodeProducer
from onnxruntime import InferenceSession
from sklearn.mixture import GaussianMixture
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
from skl2onnx import to_onnx
data = load_iris()
X_train, X_test = train_test_split(data.data)
model = GaussianMixture()
model.fit(X_train)
Default conversion¶
model_onnx = to_onnx(
model,
X_train[:1].astype(np.float32),
options={id(model): {"score_samples": True}},
target_opset=12,
)
sess = InferenceSession(
model_onnx.SerializeToString(), providers=["CPUExecutionProvider"]
)
xt = X_test[:5].astype(np.float32)
print(model.score_samples(xt))
print(sess.run(None, {"X": xt})[2])
[-3.55984426 -3.81216953 -2.14642002 -1.3580866 -2.74574493]
[[-3.559843 ]
[-3.812169 ]
[-2.1464195]
[-1.3580852]
[-2.7457438]]
Display the ONNX graph.
pydot_graph = GetPydotGraph(
model_onnx.graph,
name=model_onnx.graph.name,
rankdir="TB",
node_producer=GetOpNodeProducer(
"docstring", color="yellow", fillcolor="yellow", style="filled"
),
)
pydot_graph.write_dot("mixture.dot")
os.system("dot -O -Gdpi=300 -Tpng mixture.dot")
image = plt.imread("mixture.dot.png")
fig, ax = plt.subplots(figsize=(40, 20))
ax.imshow(image)
ax.axis("off")

(np.float64(-0.5), np.float64(4796.5), np.float64(8425.5), np.float64(-0.5))
Conversion without ReduceLogSumExp¶
Parameter black_op is used to tell the converter not to use this operator. Let’s see what the converter produces in that case.
model_onnx2 = to_onnx(
model,
X_train[:1].astype(np.float32),
options={id(model): {"score_samples": True}},
black_op={"ReduceLogSumExp"},
target_opset=12,
)
sess2 = InferenceSession(
model_onnx2.SerializeToString(), providers=["CPUExecutionProvider"]
)
xt = X_test[:5].astype(np.float32)
print(model.score_samples(xt))
print(sess2.run(None, {"X": xt})[2])
[-3.55984426 -3.81216953 -2.14642002 -1.3580866 -2.74574493]
[[-3.559843 ]
[-3.812169 ]
[-2.1464195]
[-1.3580852]
[-2.7457438]]
Display the ONNX graph.
pydot_graph = GetPydotGraph(
model_onnx2.graph,
name=model_onnx2.graph.name,
rankdir="TB",
node_producer=GetOpNodeProducer(
"docstring", color="yellow", fillcolor="yellow", style="filled"
),
)
pydot_graph.write_dot("mixture2.dot")
os.system("dot -O -Gdpi=300 -Tpng mixture2.dot")
image = plt.imread("mixture2.dot.png")
fig, ax = plt.subplots(figsize=(40, 20))
ax.imshow(image)
ax.axis("off")
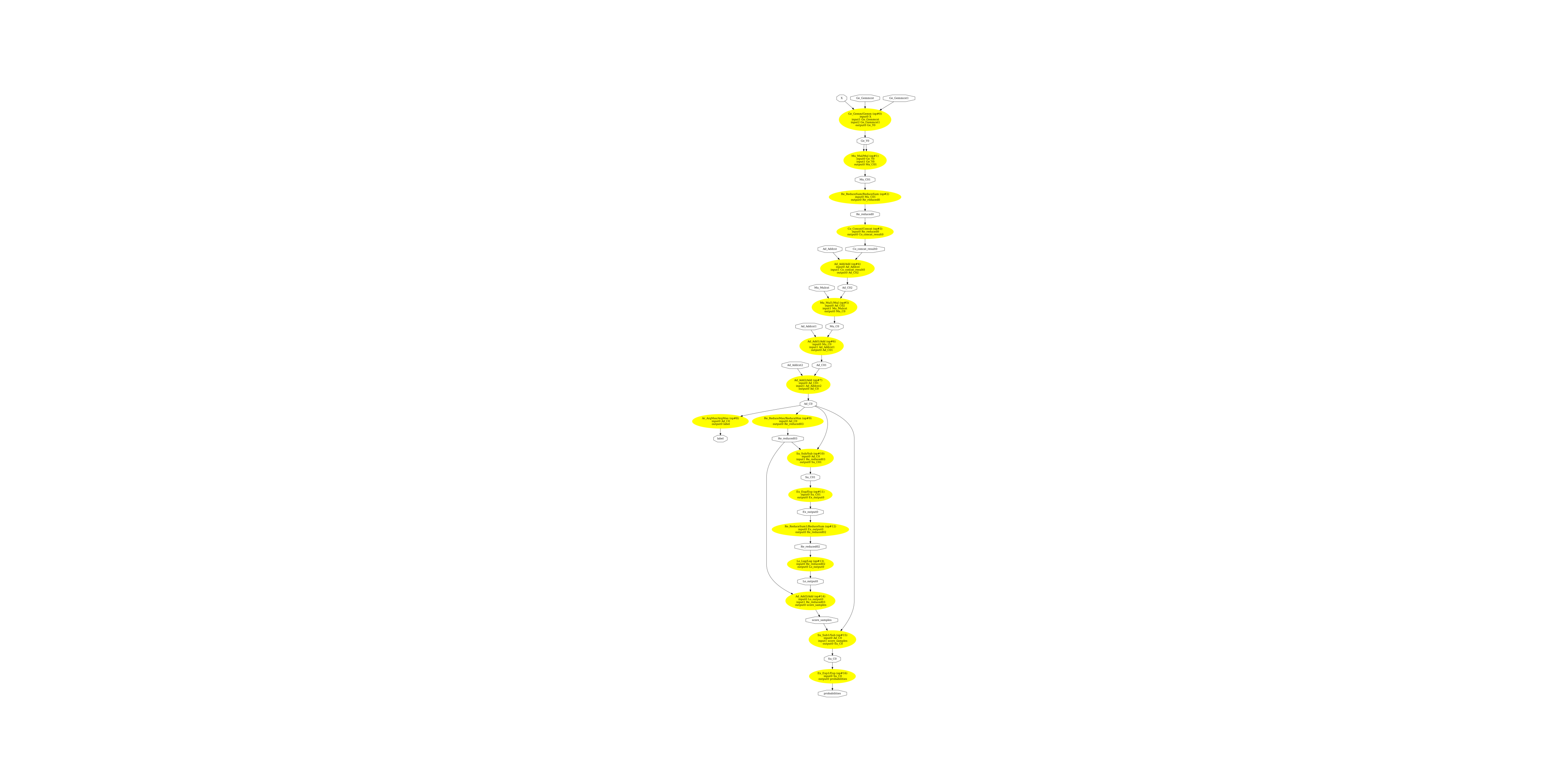
(np.float64(-0.5), np.float64(4921.5), np.float64(13264.5), np.float64(-0.5))
Processing time¶
0.2215305229999558
0.2625728419998268
The model using ReduceLogSumExp is much faster.
If the converter cannot convert without…¶
Many converters do not consider the white and black lists of operators. If a converter fails to convert without using a blacklisted operator (or only whitelisted operators), skl2onnx raises an error.
try:
to_onnx(
model,
X_train[:1].astype(np.float32),
options={id(model): {"score_samples": True}},
black_op={"ReduceLogSumExp", "Add"},
target_opset=12,
)
except RuntimeError as e:
print("Error:", e)
Error: Operator 'Add' is black listed.
Versions used for this example
import sklearn
print("numpy:", numpy.__version__)
print("scikit-learn:", sklearn.__version__)
import skl2onnx
print("onnx: ", onnx.__version__)
print("onnxruntime: ", onnxruntime.__version__)
print("skl2onnx: ", skl2onnx.__version__)
numpy: 2.3.1
scikit-learn: 1.6.1
onnx: 1.19.0
onnxruntime: 1.23.0
skl2onnx: 1.19.1
Total running time of the script: (0 minutes 17.848 seconds)