Note
Go to the end to download the full example code.
Write your own converter for your own model¶
It might happen that you implemented your own model and there is obviously no existing converter for this new model. That does not mean the conversion of a pipeline which includes it would not work. Let’s see how to do it.
t-SNE is an interesting transform which can only be used to study data as there is no way to reproduce the result once it was fitted. That’s why the class TSNE does not have any method transform, only fit_transform. This example proposes a way to train a machine learned model which approximates the outputs of a t-SNE transformer.
Implementation of the new transform¶
The first section is about the implementation. The code is quite generic but basically follows this process to fit the model with X and y:
t-SNE, \((X, y) \rightarrow X_2 \in \mathbb{R}^2\)
k nearest neightbours, \(fit(X, X_2)\), which produces function \(f(X) \rightarrow X_3\)
final normalization, simple scaling \(X_3 \rightarrow X_4\)
And to predict on a test set:
k nearest neightbours, \(f(X') \rightarrow X'_3\)
final normalization, simple scaling \(X'_3 \rightarrow X'_4\)
import inspect
import os
import numpy
import onnx
from onnx.tools.net_drawer import GetPydotGraph, GetOpNodeProducer
import onnxruntime as rt
from matplotlib import offsetbox
import matplotlib.pyplot as plt
import sklearn
from sklearn.model_selection import train_test_split
from sklearn import datasets
from sklearn.base import BaseEstimator, TransformerMixin, clone
from sklearn.manifold import TSNE
from sklearn.metrics import mean_squared_error
from sklearn.neighbors import KNeighborsRegressor
from skl2onnx import update_registered_converter
import skl2onnx
from skl2onnx import convert_sklearn, get_model_alias
from skl2onnx.common._registration import get_shape_calculator
from skl2onnx.common.data_types import FloatTensorType
class PredictableTSNE(BaseEstimator, TransformerMixin):
def __init__(
self,
transformer=None,
estimator=None,
normalize=True,
keep_tsne_outputs=False,
**kwargs,
):
"""
:param transformer: `TSNE` by default
:param estimator: `MLPRegressor` by default
:param normalize: normalizes the outputs, centers and normalizes
the output of the *t-SNE* and applies that same
normalization to he prediction of the estimator
:param keep_tsne_output: if True, keep raw outputs of
*TSNE* is stored in member *tsne_outputs_*
:param kwargs: sent to :meth:`set_params <mlinsights.mlmodel.
tsne_transformer.PredictableTSNE.set_params>`, see its
documentation to understand how to specify parameters
"""
TransformerMixin.__init__(self)
BaseEstimator.__init__(self)
if estimator is None:
estimator = KNeighborsRegressor()
if transformer is None:
transformer = TSNE()
self.estimator = estimator
self.transformer = transformer
self.keep_tsne_outputs = keep_tsne_outputs
if not hasattr(transformer, "fit_transform"):
raise AttributeError(
"Transformer {} does not have a 'fit_transform' "
"method.".format(type(transformer))
)
if not hasattr(estimator, "predict"):
raise AttributeError(
"Estimator {} does not have a 'predict' method.".format(type(estimator))
)
self.normalize = normalize
if kwargs:
self.set_params(**kwargs)
def fit(self, X, y, sample_weight=None):
"""
Runs a *k-means* on each class
then trains a classifier on the
extended set of features.
Parameters
----------
X : numpy array or sparse matrix of shape [n_samples,n_features]
Training data
y : numpy array of shape [n_samples, n_targets]
Target values. Will be cast to X's dtype if necessary
sample_weight : numpy array of shape [n_samples]
Individual weights for each sample
Returns
-------
self : returns an instance of self.
Attributes
----------
transformer_: trained transformeer
estimator_: trained regressor
tsne_outputs_: t-SNE outputs if *keep_tsne_outputs* is True
mean_: average of the *t-SNE* output on each dimension
inv_std_: inverse of the standard deviation of the *t-SNE*
output on each dimension
loss_: loss (*mean_squared_error*)
between the predictions and the outputs of t-SNE
"""
params = dict(y=y, sample_weight=sample_weight)
self.transformer_ = clone(self.transformer)
sig = inspect.signature(self.transformer.fit_transform)
pars = {}
for p in ["sample_weight", "y"]:
if p in sig.parameters and p in params:
pars[p] = params[p]
target = self.transformer_.fit_transform(X, **pars)
sig = inspect.signature(self.estimator.fit)
if "sample_weight" in sig.parameters:
self.estimator_ = clone(self.estimator).fit(
X, target, sample_weight=sample_weight
)
else:
self.estimator_ = clone(self.estimator).fit(X, target)
mean = target.mean(axis=0)
var = target.std(axis=0)
self.mean_ = mean
self.inv_std_ = 1.0 / var
exp = (target - mean) * self.inv_std_
got = (self.estimator_.predict(X) - mean) * self.inv_std_
self.loss_ = mean_squared_error(exp, got)
if self.keep_tsne_outputs:
self.tsne_outputs_ = exp if self.normalize else target
return self
def transform(self, X):
"""
Runs the predictions.
Parameters
----------
X : numpy array or sparse matrix of shape [n_samples,n_features]
Training data
Returns
-------
tranformed *X*
"""
pred = self.estimator_.predict(X)
if self.normalize:
pred -= self.mean_
pred *= self.inv_std_
return pred
def get_params(self, deep=True):
"""
Returns the parameters for all the embedded objects.
"""
res = {}
for k, v in self.transformer.get_params().items():
res["t_" + k] = v
for k, v in self.estimator.get_params().items():
res["e_" + k] = v
return res
def set_params(self, **values):
"""
Sets the parameters before training.
Every parameter prefixed by ``'e_'`` is an estimator
parameter, every parameter prefixed by
``t_`` is for a transformer parameter.
"""
pt, pe, pn = {}, {}, {}
for k, v in values.items():
if k.startswith("e_"):
pe[k[2:]] = v
elif k.startswith("t_"):
pt[k[2:]] = v
elif k.startswith("n_"):
pn[k[2:]] = v
else:
raise ValueError("Unexpected parameter name '{0}'.".format(k))
self.transformer.set_params(**pt)
self.estimator.set_params(**pe)
Experimentation on MNIST¶
Let’s fit t-SNE…
digits = datasets.load_digits(n_class=6)
Xd = digits.data
yd = digits.target
imgs = digits.images
n_samples, n_features = Xd.shape
n_samples, n_features
X_train, X_test, y_train, y_test, imgs_train, imgs_test = train_test_split(Xd, yd, imgs)
tsne = TSNE(n_components=2, init="pca", random_state=0)
def plot_embedding(Xp, y, imgs, title=None, figsize=(12, 4)):
x_min, x_max = numpy.min(Xp, 0), numpy.max(Xp, 0)
X = (Xp - x_min) / (x_max - x_min)
_fig, ax = plt.subplots(1, 2, figsize=figsize)
for i in range(X.shape[0]):
ax[0].text(
X[i, 0],
X[i, 1],
str(y[i]),
color=plt.cm.Set1(y[i] / 10.0),
fontdict={"weight": "bold", "size": 9},
)
if hasattr(offsetbox, "AnnotationBbox"):
# only print thumbnails with matplotlib > 1.0
shown_images = numpy.array([[1.0, 1.0]]) # just something big
for i in range(X.shape[0]):
dist = numpy.sum((X[i] - shown_images) ** 2, 1)
if numpy.min(dist) < 4e-3:
# don't show points that are too close
continue
shown_images = numpy.r_[shown_images, [X[i]]]
imagebox = offsetbox.AnnotationBbox(
offsetbox.OffsetImage(imgs[i], cmap=plt.cm.gray_r), X[i]
)
ax[0].add_artist(imagebox)
ax[0].set_xticks([]), ax[0].set_yticks([])
ax[1].plot(Xp[:, 0], Xp[:, 1], ".")
if title is not None:
ax[0].set_title(title)
return ax
X_train_tsne = tsne.fit_transform(X_train)
plot_embedding(X_train_tsne, y_train, imgs_train, "t-SNE embedding of the digits")
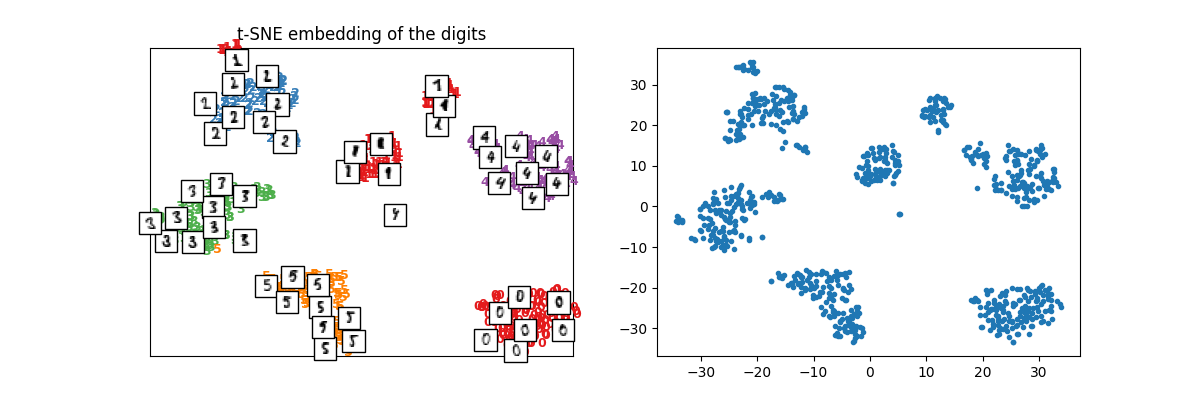
array([<Axes: title={'center': 't-SNE embedding of the digits'}>,
<Axes: >], dtype=object)
Repeatable t-SNE¶
Just to check it is working.
ptsne_knn = PredictableTSNE()
ptsne_knn.fit(X_train, y_train)
X_train_tsne2 = ptsne_knn.transform(X_train)
plot_embedding(
X_train_tsne2,
y_train,
imgs_train,
"Predictable t-SNE of the digits\nStandardScaler+KNeighborsRegressor",
)
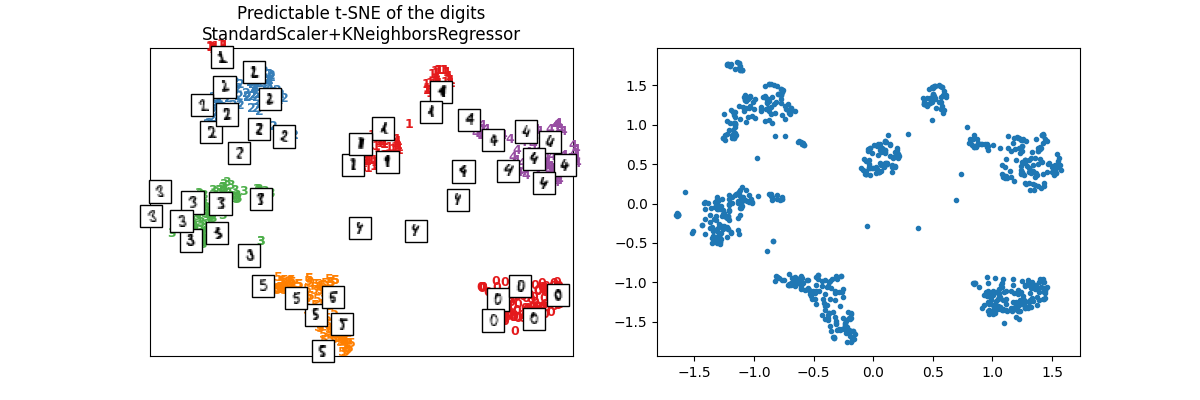
array([<Axes: title={'center': 'Predictable t-SNE of the digits\nStandardScaler+KNeighborsRegressor'}>,
<Axes: >], dtype=object)
We check on test set.
X_test_tsne2 = ptsne_knn.transform(X_test)
plot_embedding(
X_test_tsne2,
y_test,
imgs_test,
"Predictable t-SNE of the digits\nStandardScaler+KNeighborsRegressor",
)
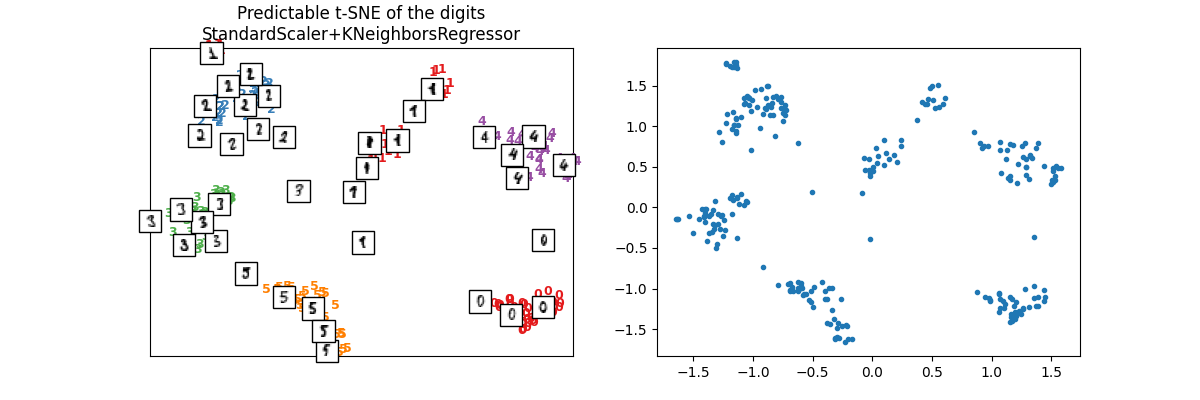
array([<Axes: title={'center': 'Predictable t-SNE of the digits\nStandardScaler+KNeighborsRegressor'}>,
<Axes: >], dtype=object)
ONNX - shape_calculator, converter¶
Now starts the part dedicated to ONNX. ONNX conversion requires two function, one to calculate the shape of the outputs based on the inputs, the other one to do the actual conversion of the model.
def predictable_tsne_shape_calculator(operator):
input = operator.inputs[0] # inputs in ONNX graph
# output = operator.outputs[0] # output in ONNX graph
op = operator.raw_operator # scikit-learn model (mmust be fitted)
N = input.type.shape[0] # number of observations
C = op.estimator_._y.shape[1] # dimension of outputs
# new output definition
operator.outputs[0].type = FloatTensorType([N, C])
Then the converter model. We reuse existing converter.
def predictable_tsne_converter(scope, operator, container):
"""
:param scope: name space, where to keep node names, get unused new names
:param operator: operator to converter, same object as sent to
*predictable_tsne_shape_calculator*
:param container: contains the ONNX graph
"""
# input = operator.inputs[0] # input in ONNX graph
output = operator.outputs[0] # output in ONNX graph
op = operator.raw_operator # scikit-learn model (mmust be fitted)
# First step is the k nearest-neighbours,
# we reuse existing converter and declare it as local
# operator.
model = op.estimator_
alias = get_model_alias(type(model))
knn_op = scope.declare_local_operator(alias, model)
knn_op.inputs = operator.inputs
# We add an intermediate outputs.
knn_output = scope.declare_local_variable("knn_output", FloatTensorType())
knn_op.outputs.append(knn_output)
# We adjust the output of the submodel.
shape_calc = get_shape_calculator(alias)
shape_calc(knn_op)
# We add the normalizer which needs a unique node name.
name = scope.get_unique_operator_name("Scaler")
# The parameter follows the specifications of ONNX
# https://github.com/onnx/onnx/blob/main/docs/Operators-ml.md#ai.onnx.ml.Scaler
attrs = dict(
name=name,
scale=op.inv_std_.ravel().astype(numpy.float32),
offset=op.mean_.ravel().astype(numpy.float32),
)
# Let's finally add the scaler which connects the output
# of the k-nearest neighbours model to output of the whole model
# declared in ONNX graph
container.add_node(
"Scaler",
[knn_output.onnx_name],
[output.full_name],
op_domain="ai.onnx.ml",
**attrs,
)
We now need to declare the new converter.
update_registered_converter(
PredictableTSNE,
"CustomPredictableTSNE",
predictable_tsne_shape_calculator,
predictable_tsne_converter,
)
Conversion to ONNX¶
We just need to call convert_sklearn as any other model to convert.
model_onnx = convert_sklearn(
ptsne_knn,
"predictable_tsne",
[("input", FloatTensorType([None, X_test.shape[1]]))],
target_opset=12,
)
# And save.
with open("predictable_tsne.onnx", "wb") as f:
f.write(model_onnx.SerializeToString())
We now compare the prediction.
print("ptsne_knn.tranform\n", ptsne_knn.transform(X_test[:2]))
ptsne_knn.tranform
[[ 0.1792845 0.7067502 ]
[-0.27657354 -0.9149268 ]]
Predictions with onnxruntime.
sess = rt.InferenceSession("predictable_tsne.onnx", providers=["CPUExecutionProvider"])
pred_onx = sess.run(None, {"input": X_test[:1].astype(numpy.float32)})
print("transform", pred_onx[0])
transform [[0.1792845 0.7067502]]
The converter for the nearest neighbours produces an ONNX graph which does not allow multiple predictions at a time. Let’s call onnxruntime for the second row.
pred_onx = sess.run(None, {"input": X_test[1:2].astype(numpy.float32)})
print("transform", pred_onx[0])
transform [[-0.27657354 -0.9149268 ]]
Display the ONNX graph¶
pydot_graph = GetPydotGraph(
model_onnx.graph,
name=model_onnx.graph.name,
rankdir="TB",
node_producer=GetOpNodeProducer(
"docstring", color="yellow", fillcolor="yellow", style="filled"
),
)
pydot_graph.write_dot("pipeline_tsne.dot")
os.system("dot -O -Gdpi=300 -Tpng pipeline_tsne.dot")
image = plt.imread("pipeline_tsne.dot.png")
fig, ax = plt.subplots(figsize=(40, 20))
ax.imshow(image)
ax.axis("off")

(np.float64(-0.5), np.float64(2643.5), np.float64(9099.5), np.float64(-0.5))
Versions used for this example
print("numpy:", numpy.__version__)
print("scikit-learn:", sklearn.__version__)
print("onnx: ", onnx.__version__)
print("onnxruntime: ", rt.__version__)
print("skl2onnx: ", skl2onnx.__version__)
numpy: 2.4.1
scikit-learn: 1.8.0
onnx: 1.21.0
onnxruntime: 1.24.0
skl2onnx: 1.20.0
Total running time of the script: (0 minutes 7.884 seconds)