Note
Go to the end to download the full example code.
When a custom model is neither a classifier nor a regressor¶
scikit-learn’s API specifies that a regressor produces one outputs and a classifier produces two outputs, predicted labels and probabilities. The goal here is to add a third result which tells if the probability is above a given threshold. That’s implemented in method validate.
Iris and scoring¶
A new class is created, it trains any classifier and implements the method validate mentioned above.
import inspect
import numpy as np
import skl2onnx
import onnx
import sklearn
from sklearn.base import ClassifierMixin, BaseEstimator, clone
from sklearn.datasets import load_iris
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from skl2onnx import update_registered_converter
import os
from onnx.tools.net_drawer import GetPydotGraph, GetOpNodeProducer
import onnxruntime as rt
from skl2onnx.common._apply_operation import apply_identity, apply_cast, apply_greater
from skl2onnx import to_onnx, get_model_alias
from skl2onnx.proto import onnx_proto
from skl2onnx.common._registration import get_shape_calculator
from skl2onnx.common.data_types import FloatTensorType, Int64TensorType
import matplotlib.pyplot as plt
class ValidatorClassifier(BaseEstimator, ClassifierMixin):
def __init__(self, estimator=None, threshold=0.75):
ClassifierMixin.__init__(self)
BaseEstimator.__init__(self)
if estimator is None:
estimator = LogisticRegression()
self.estimator = estimator
self.threshold = threshold
def fit(self, X, y, sample_weight=None):
sig = inspect.signature(self.estimator.fit)
if "sample_weight" in sig.parameters:
self.estimator_ = clone(self.estimator).fit(
X, y, sample_weight=sample_weight
)
else:
self.estimator_ = clone(self.estimator).fit(X, y)
return self
def predict(self, X):
return self.estimator_.predict(X)
def predict_proba(self, X):
return self.estimator_.predict_proba(X)
def validate(self, X):
pred = self.predict_proba(X)
mx = pred.max(axis=1)
return (mx >= self.threshold) * 1
data = load_iris()
X, y = data.data, data.target
X_train, X_test, y_train, y_test = train_test_split(X, y)
model = ValidatorClassifier()
model.fit(X_train, y_train)
Let’s now measure the indicator which tells if the probability of a prediction is above a threshold.
print(model.validate(X_test))
[1 1 1 1 1 1 1 1 1 1 0 1 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 1 1 1 1 1 1 0
1]
Conversion to ONNX¶
The conversion fails for a new model because the library does not know any converter associated to this new model.
try:
to_onnx(model, X_train[:1].astype(np.float32), target_opset=12)
except RuntimeError as e:
print(e)
Unable to find a shape calculator for type '<class '__main__.ValidatorClassifier'>'.
It usually means the pipeline being converted contains a
transformer or a predictor with no corresponding converter
implemented in sklearn-onnx. If the converted is implemented
in another library, you need to register
the converted so that it can be used by sklearn-onnx (function
update_registered_converter). If the model is not yet covered
by sklearn-onnx, you may raise an issue to
https://github.com/onnx/sklearn-onnx/issues
to get the converter implemented or even contribute to the
project. If the model is a custom model, a new converter must
be implemented. Examples can be found in the gallery.
Custom converter¶
We reuse some pieces of code from Write your own converter for your own model. The shape calculator defines the shape of every output of the converted model.
def validator_classifier_shape_calculator(operator):
input0 = operator.inputs[0] # inputs in ONNX graph
outputs = operator.outputs # outputs in ONNX graph
op = operator.raw_operator # scikit-learn model (mmust be fitted)
if len(outputs) != 3:
raise RuntimeError("3 outputs expected not {}.".format(len(outputs)))
N = input0.type.shape[0] # number of observations
C = op.estimator_.classes_.shape[0] # dimension of outputs
outputs[0].type = Int64TensorType([N]) # label
outputs[1].type = FloatTensorType([N, C]) # probabilities
outputs[2].type = Int64TensorType([C]) # validation
Then the converter.
def validator_classifier_converter(scope, operator, container):
outputs = operator.outputs # outputs in ONNX graph
op = operator.raw_operator # scikit-learn model (mmust be fitted)
# We reuse existing converter and declare it
# as a local operator.
model = op.estimator_
alias = get_model_alias(type(model))
val_op = scope.declare_local_operator(alias, model)
val_op.inputs = operator.inputs
# We add an intermediate outputs.
val_label = scope.declare_local_variable("val_label", Int64TensorType())
val_prob = scope.declare_local_variable("val_prob", FloatTensorType())
val_op.outputs.append(val_label)
val_op.outputs.append(val_prob)
# We adjust the output of the submodel.
shape_calc = get_shape_calculator(alias)
shape_calc(val_op)
# We now handle the validation.
val_max = scope.get_unique_variable_name("val_max")
if container.target_opset >= 18:
axis_name = scope.get_unique_variable_name("axis")
container.add_initializer(axis_name, onnx_proto.TensorProto.INT64, [1], [1])
container.add_node(
"ReduceMax",
[val_prob.full_name, axis_name],
val_max,
name=scope.get_unique_operator_name("ReduceMax"),
keepdims=0,
)
else:
container.add_node(
"ReduceMax",
val_prob.full_name,
val_max,
name=scope.get_unique_operator_name("ReduceMax"),
axes=[1],
keepdims=0,
)
th_name = scope.get_unique_variable_name("threshold")
container.add_initializer(
th_name, onnx_proto.TensorProto.FLOAT, [1], [op.threshold]
)
val_bin = scope.get_unique_variable_name("val_bin")
apply_greater(scope, [val_max, th_name], val_bin, container)
val_val = scope.get_unique_variable_name("validate")
apply_cast(scope, val_bin, val_val, container, to=onnx_proto.TensorProto.INT64)
# We finally link the intermediate output to the shared converter.
apply_identity(scope, val_label.full_name, outputs[0].full_name, container)
apply_identity(scope, val_prob.full_name, outputs[1].full_name, container)
apply_identity(scope, val_val, outputs[2].full_name, container)
Then the registration.
update_registered_converter(
ValidatorClassifier,
"CustomValidatorClassifier",
validator_classifier_shape_calculator,
validator_classifier_converter,
)
And conversion…
try:
to_onnx(model, X_test[:1].astype(np.float32), target_opset=12)
except RuntimeError as e:
print(e)
3 outputs expected not 2.
It fails because the library expected the model to behave like a classifier which produces two outputs. We need to add a custom parser to tell the library this model produces three outputs.
Custom parser¶
def validator_classifier_parser(scope, model, inputs, custom_parsers=None):
alias = get_model_alias(type(model))
this_operator = scope.declare_local_operator(alias, model)
# inputs
this_operator.inputs.append(inputs[0])
# outputs
val_label = scope.declare_local_variable("val_label", Int64TensorType())
val_prob = scope.declare_local_variable("val_prob", FloatTensorType())
val_val = scope.declare_local_variable("val_val", Int64TensorType())
this_operator.outputs.append(val_label)
this_operator.outputs.append(val_prob)
this_operator.outputs.append(val_val)
# end
return this_operator.outputs
Registration.
update_registered_converter(
ValidatorClassifier,
"CustomValidatorClassifier",
validator_classifier_shape_calculator,
validator_classifier_converter,
parser=validator_classifier_parser,
)
And conversion again.
model_onnx = to_onnx(model, X_test[:1].astype(np.float32), target_opset=12)
Final test¶
We need now to check the results are the same with ONNX.
X32 = X_test[:5].astype(np.float32)
sess = rt.InferenceSession(
model_onnx.SerializeToString(), providers=["CPUExecutionProvider"]
)
results = sess.run(None, {"X": X32})
print("--labels--")
print("sklearn", model.predict(X32))
print("onnx", results[0])
print("--probabilities--")
print("sklearn", model.predict_proba(X32))
print("onnx", results[1])
print("--validation--")
print("sklearn", model.validate(X32))
print("onnx", results[2])
--labels--
sklearn [0 2 2 2 1]
onnx [0 2 2 2 1]
--probabilities--
sklearn [[9.43060711e-01 5.69381890e-02 1.09973143e-06]
[2.49010751e-05 1.99003237e-02 9.80074775e-01]
[1.95285668e-06 2.67317977e-02 9.73266249e-01]
[3.00896099e-07 4.43030316e-03 9.95569396e-01]
[1.25373515e-02 9.66555985e-01 2.09066639e-02]]
onnx [[9.4306076e-01 5.6938179e-02 1.0997300e-06]
[2.4901081e-05 1.9900326e-02 9.8007476e-01]
[1.9528586e-06 2.6731813e-02 9.7326624e-01]
[3.0089643e-07 4.4303052e-03 9.9556941e-01]
[1.2537363e-02 9.6655601e-01 2.0906653e-02]]
--validation--
sklearn [1 1 1 1 1]
onnx [1 1 1 1 1]
It looks good.
Display the ONNX graph¶
pydot_graph = GetPydotGraph(
model_onnx.graph,
name=model_onnx.graph.name,
rankdir="TB",
node_producer=GetOpNodeProducer(
"docstring", color="yellow", fillcolor="yellow", style="filled"
),
)
pydot_graph.write_dot("validator_classifier.dot")
os.system("dot -O -Gdpi=300 -Tpng validator_classifier.dot")
image = plt.imread("validator_classifier.dot.png")
fig, ax = plt.subplots(figsize=(40, 20))
ax.imshow(image)
ax.axis("off")
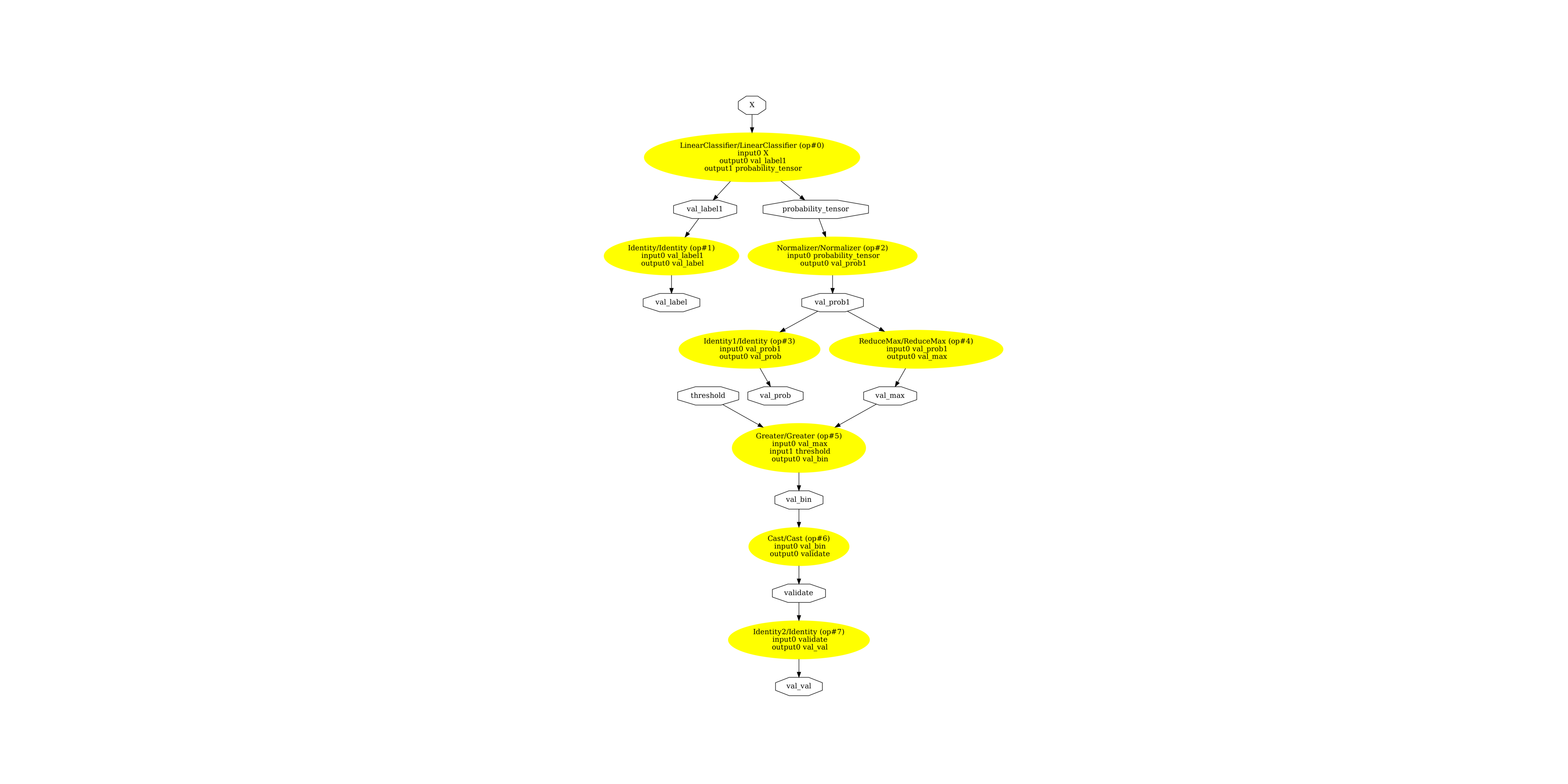
(np.float64(-0.5), np.float64(3293.5), np.float64(4934.5), np.float64(-0.5))
Versions used for this example
print("numpy:", np.__version__)
print("scikit-learn:", sklearn.__version__)
print("onnx: ", onnx.__version__)
print("onnxruntime: ", rt.__version__)
print("skl2onnx: ", skl2onnx.__version__)
numpy: 2.4.1
scikit-learn: 1.8.0
onnx: 1.21.0
onnxruntime: 1.24.0
skl2onnx: 1.20.0
Total running time of the script: (0 minutes 2.675 seconds)