Note
Go to the end to download the full example code.
Play with ONNX operators¶
ONNX aims at describing most of the machine learning models implemented in scikit-learn but it does not necessarily describe the prediction function the same way scikit-learn does. If it is possible to define custom operators, it usually requires some time to add it to ONNX specifications and then to the backend used to compute the predictions. It is better to look first if the existing operators can be used. The list is available on github and gives the basic operators and others dedicated to machine learning. ONNX has a Python API which can be used to define an ONNX graph: PythonAPIOverview.md. But it is quite verbose and makes it difficult to describe big graphs. sklearn-onnx implements a nicer way to test ONNX operators.
ONNX Python API¶
Let’s try the example given by ONNX documentation: ONNX Model Using Helper Functions. It relies on protobuf whose definition can be found on github onnx.proto.
import onnxruntime
import numpy
import os
import numpy as np
import matplotlib.pyplot as plt
import onnx
from onnx import helper
from onnx import TensorProto
from onnx.tools.net_drawer import GetPydotGraph, GetOpNodeProducer
# Create one input (ValueInfoProto)
X = helper.make_tensor_value_info("X", TensorProto.FLOAT, [None, 2])
# Create one output (ValueInfoProto)
Y = helper.make_tensor_value_info("Y", TensorProto.FLOAT, [None, 4])
# Create a node (NodeProto)
node_def = helper.make_node(
"Pad", # node name
["X"], # inputs
["Y"], # outputs
mode="constant", # attributes
value=1.5,
pads=[0, 1, 0, 1],
)
# Create the graph (GraphProto)
graph_def = helper.make_graph(
[node_def],
"test-model",
[X],
[Y],
)
# Create the model (ModelProto)
model_def = helper.make_model(graph_def, producer_name="onnx-example")
model_def.opset_import[0].version = 10
print("The model is:\n{}".format(model_def))
onnx.checker.check_model(model_def)
print("The model is checked!")
The model is:
ir_version: 13
producer_name: "onnx-example"
graph {
node {
input: "X"
output: "Y"
op_type: "Pad"
attribute {
name: "mode"
s: "constant"
type: STRING
}
attribute {
name: "pads"
ints: 0
ints: 1
ints: 0
ints: 1
type: INTS
}
attribute {
name: "value"
f: 1.5
type: FLOAT
}
}
name: "test-model"
input {
name: "X"
type {
tensor_type {
elem_type: 1
shape {
dim {
}
dim {
dim_value: 2
}
}
}
}
}
output {
name: "Y"
type {
tensor_type {
elem_type: 1
shape {
dim {
}
dim {
dim_value: 4
}
}
}
}
}
}
opset_import {
version: 10
}
The model is checked!
Same example with sklearn-onnx¶
Every operator has its own class in sklearn-onnx. The list is dynamically created based on the installed onnx package.
from skl2onnx.algebra.onnx_ops import OnnxPad
pad = OnnxPad(
"X",
output_names=["Y"],
mode="constant",
value=1.5,
pads=[0, 1, 0, 1],
op_version=10,
)
model_def = pad.to_onnx({"X": X}, target_opset=10)
print("The model is:\n{}".format(model_def))
onnx.checker.check_model(model_def)
print("The model is checked!")
The model is:
ir_version: 5
producer_name: "skl2onnx"
producer_version: "1.20.0"
domain: "ai.onnx"
model_version: 0
graph {
node {
input: "X"
output: "Y"
name: "Pa_Pad"
op_type: "Pad"
attribute {
name: "mode"
s: "constant"
type: STRING
}
attribute {
name: "pads"
ints: 0
ints: 1
ints: 0
ints: 1
type: INTS
}
attribute {
name: "value"
f: 1.5
type: FLOAT
}
domain: ""
}
name: "OnnxPad"
input {
name: "X"
type {
tensor_type {
elem_type: 1
shape {
dim {
}
dim {
dim_value: 2
}
}
}
}
}
output {
name: "Y"
type {
tensor_type {
elem_type: 1
shape {
dim {
}
dim {
dim_value: 4
}
}
}
}
}
}
opset_import {
domain: ""
version: 10
}
The model is checked!
Inputs and outputs can also be skipped.
pad = OnnxPad(mode="constant", value=1.5, pads=[0, 1, 0, 1], op_version=10)
model_def = pad.to_onnx({pad.inputs[0].name: X}, target_opset=10)
onnx.checker.check_model(model_def)
Multiple operators¶
Let’s use the second example from the documentation.
# Preprocessing: create a model with two nodes, Y's shape is unknown
node1 = helper.make_node("Transpose", ["X"], ["Y"], perm=[1, 0, 2])
node2 = helper.make_node("Transpose", ["Y"], ["Z"], perm=[1, 0, 2])
graph = helper.make_graph(
[node1, node2],
"two-transposes",
[helper.make_tensor_value_info("X", TensorProto.FLOAT, (2, 3, 4))],
[helper.make_tensor_value_info("Z", TensorProto.FLOAT, (2, 3, 4))],
)
original_model = helper.make_model(graph, producer_name="onnx-examples")
# Check the model and print Y's shape information
onnx.checker.check_model(original_model)
Which we translate into:
from skl2onnx.algebra.onnx_ops import OnnxTranspose
node = OnnxTranspose(
OnnxTranspose("X", perm=[1, 0, 2], op_version=12), perm=[1, 0, 2], op_version=12
)
X = np.arange(2 * 3 * 4).reshape((2, 3, 4)).astype(np.float32)
# numpy arrays are good enough to define the input shape
model_def = node.to_onnx({"X": X}, target_opset=12)
onnx.checker.check_model(model_def)
Let’s the output with onnxruntime
def predict_with_onnxruntime(model_def, *inputs):
import onnxruntime as ort
sess = ort.InferenceSession(
model_def.SerializeToString(), providers=["CPUExecutionProvider"]
)
names = [i.name for i in sess.get_inputs()]
dinputs = dict(zip(names, inputs))
res = sess.run(None, dinputs)
names = [o.name for o in sess.get_outputs()]
return dict(zip(names, res))
Y = predict_with_onnxruntime(model_def, X)
print(Y)
{'Tr_transposed0': array([[[ 0., 1., 2., 3.],
[ 4., 5., 6., 7.],
[ 8., 9., 10., 11.]],
[[12., 13., 14., 15.],
[16., 17., 18., 19.],
[20., 21., 22., 23.]]], dtype=float32)}
Display the ONNX graph¶
pydot_graph = GetPydotGraph(
model_def.graph,
name=model_def.graph.name,
rankdir="TB",
node_producer=GetOpNodeProducer(
"docstring", color="yellow", fillcolor="yellow", style="filled"
),
)
pydot_graph.write_dot("pipeline_transpose2x.dot")
os.system("dot -O -Gdpi=300 -Tpng pipeline_transpose2x.dot")
image = plt.imread("pipeline_transpose2x.dot.png")
fig, ax = plt.subplots(figsize=(40, 20))
ax.imshow(image)
ax.axis("off")
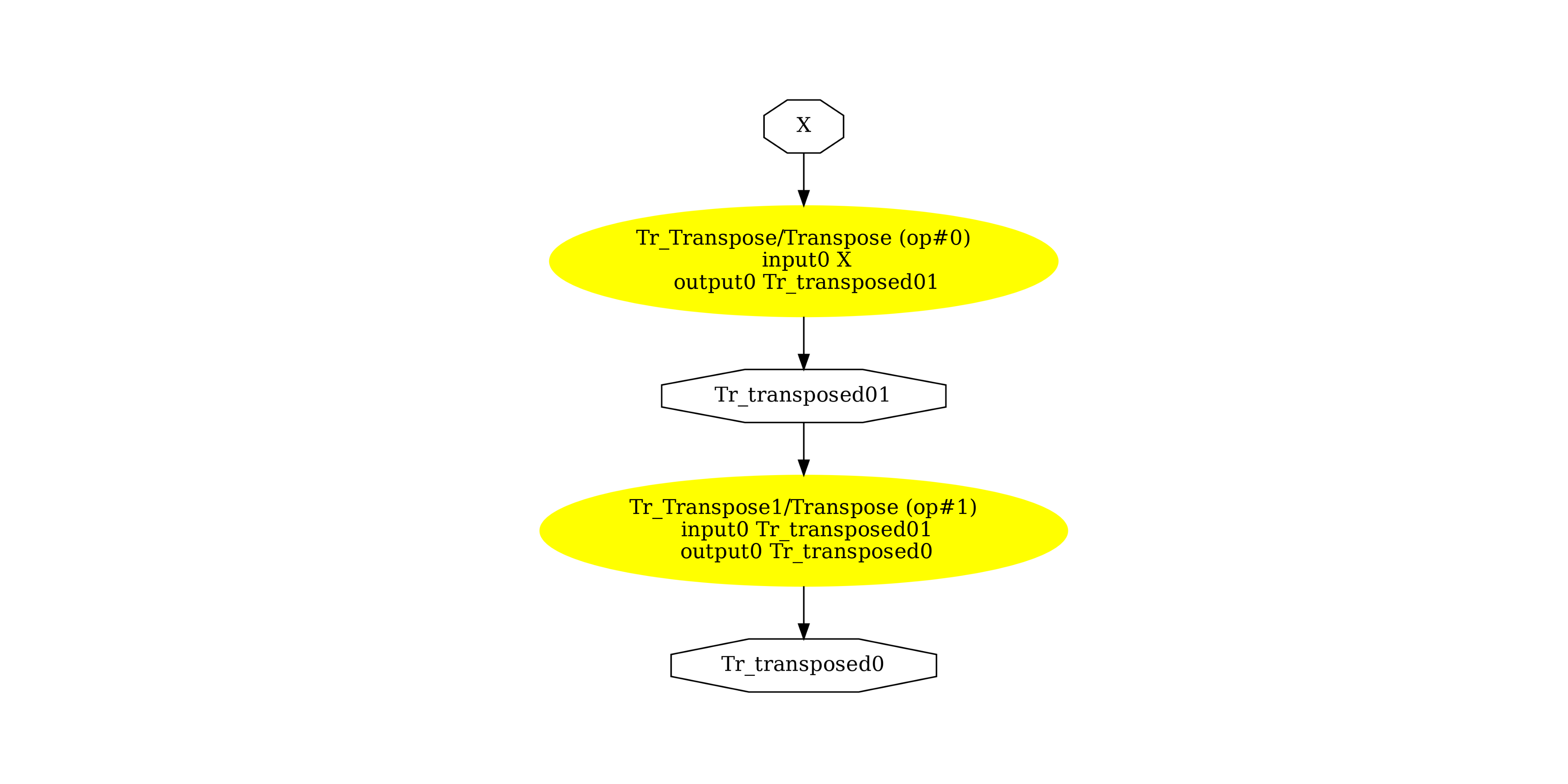
(np.float64(-0.5), np.float64(1524.5), np.float64(1707.5), np.float64(-0.5))
Versions used for this example
import sklearn
print("numpy:", numpy.__version__)
print("scikit-learn:", sklearn.__version__)
import skl2onnx
print("onnx: ", onnx.__version__)
print("onnxruntime: ", onnxruntime.__version__)
print("skl2onnx: ", skl2onnx.__version__)
numpy: 2.4.1
scikit-learn: 1.8.0
onnx: 1.21.0
onnxruntime: 1.24.0
skl2onnx: 1.20.0
Total running time of the script: (0 minutes 0.702 seconds)