Note
Go to the end to download the full example code.
Benchmark ONNX conversion¶
Example Train and deploy a scikit-learn pipeline converts a simple model. This example takes a similar example but on random data and compares the processing time required by each option to compute predictions.
Training a pipeline¶
import numpy
from pandas import DataFrame
from tqdm import tqdm
from onnx.reference import ReferenceEvaluator
from sklearn import config_context
from sklearn.datasets import make_regression
from sklearn.ensemble import (
GradientBoostingRegressor,
RandomForestRegressor,
VotingRegressor,
)
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
from onnxruntime import InferenceSession
from skl2onnx import to_onnx
from skl2onnx.tutorial import measure_time
N = 11000
X, y = make_regression(N, n_features=10)
X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=0.01)
print("Train shape", X_train.shape)
print("Test shape", X_test.shape)
reg1 = GradientBoostingRegressor(random_state=1)
reg2 = RandomForestRegressor(random_state=1)
reg3 = LinearRegression()
ereg = VotingRegressor([("gb", reg1), ("rf", reg2), ("lr", reg3)])
ereg.fit(X_train, y_train)
Train shape (110, 10)
Test shape (10890, 10)
Measure the processing time¶
We use function skl2onnx.tutorial.measure_time().
The page about assume_finite
may be useful if you need to optimize the prediction.
We measure the processing time per observation whether
or not an observation belongs to a batch or is a single one.
sizes = [(1, 50), (10, 50), (100, 10)]
with config_context(assume_finite=True):
obs = []
for batch_size, repeat in tqdm(sizes):
context = {"ereg": ereg, "X": X_test[:batch_size]}
mt = measure_time(
"ereg.predict(X)", context, div_by_number=True, number=10, repeat=repeat
)
mt["size"] = context["X"].shape[0]
mt["mean_obs"] = mt["average"] / mt["size"]
obs.append(mt)
df_skl = DataFrame(obs)
df_skl
0%| | 0/3 [00:00<?, ?it/s]
33%|███▎ | 1/3 [00:04<00:08, 4.01s/it]
67%|██████▋ | 2/3 [00:07<00:03, 3.74s/it]
100%|██████████| 3/3 [00:08<00:00, 2.44s/it]
100%|██████████| 3/3 [00:08<00:00, 2.82s/it]
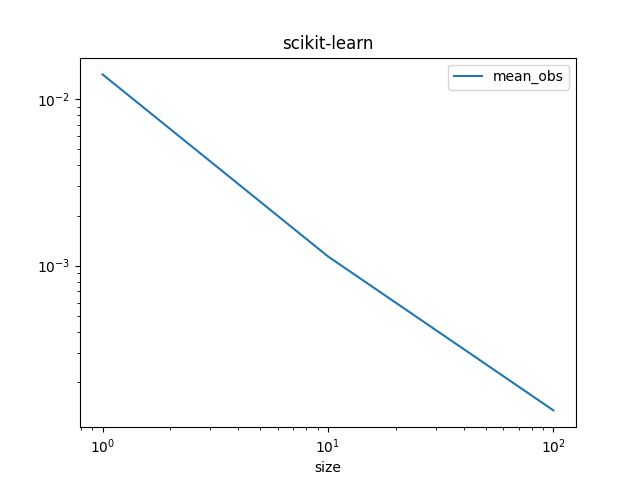
Graphe.
df_skl.set_index("size")[["mean_obs"]].plot(title="scikit-learn", logx=True, logy=True)
ONNX runtime¶
The same is done with the two ONNX runtime available.
onx = to_onnx(ereg, X_train[:1].astype(numpy.float32), target_opset=14)
sess = InferenceSession(onx.SerializeToString(), providers=["CPUExecutionProvider"])
oinf = ReferenceEvaluator(onx)
obs = []
for batch_size, repeat in tqdm(sizes):
# scikit-learn
context = {"ereg": ereg, "X": X_test[:batch_size].astype(numpy.float32)}
mt = measure_time(
"ereg.predict(X)", context, div_by_number=True, number=10, repeat=repeat
)
mt["size"] = context["X"].shape[0]
mt["skl"] = mt["average"] / mt["size"]
# onnxruntime
context = {"sess": sess, "X": X_test[:batch_size].astype(numpy.float32)}
mt2 = measure_time(
"sess.run(None, {'X': X})[0]",
context,
div_by_number=True,
number=10,
repeat=repeat,
)
mt["ort"] = mt2["average"] / mt["size"]
# ReferenceEvaluator
context = {"oinf": oinf, "X": X_test[:batch_size].astype(numpy.float32)}
mt2 = measure_time(
"oinf.run(None, {'X': X})[0]",
context,
div_by_number=True,
number=10,
repeat=repeat,
)
mt["pyrt"] = mt2["average"] / mt["size"]
# end
obs.append(mt)
df = DataFrame(obs)
df
0%| | 0/3 [00:00<?, ?it/s]
33%|███▎ | 1/3 [00:09<00:18, 9.12s/it]
67%|██████▋ | 2/3 [00:24<00:12, 12.77s/it]
100%|██████████| 3/3 [00:38<00:00, 13.40s/it]
100%|██████████| 3/3 [00:38<00:00, 12.86s/it]
Graph.
df.set_index("size")[["skl", "ort", "pyrt"]].plot(
title="Average prediction time per runtime", logx=True, logy=True
)
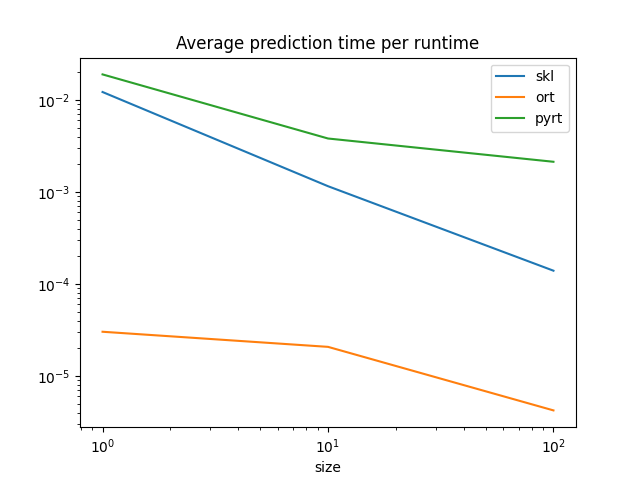
ONNX runtimes are much faster than scikit-learn to predict one observation. scikit-learn is optimized for training, for batch prediction. That explains why scikit-learn and ONNX runtimes seem to converge for big batches. They use similar implementation, parallelization and languages (C++, openmp).
Total running time of the script: (0 minutes 47.782 seconds)